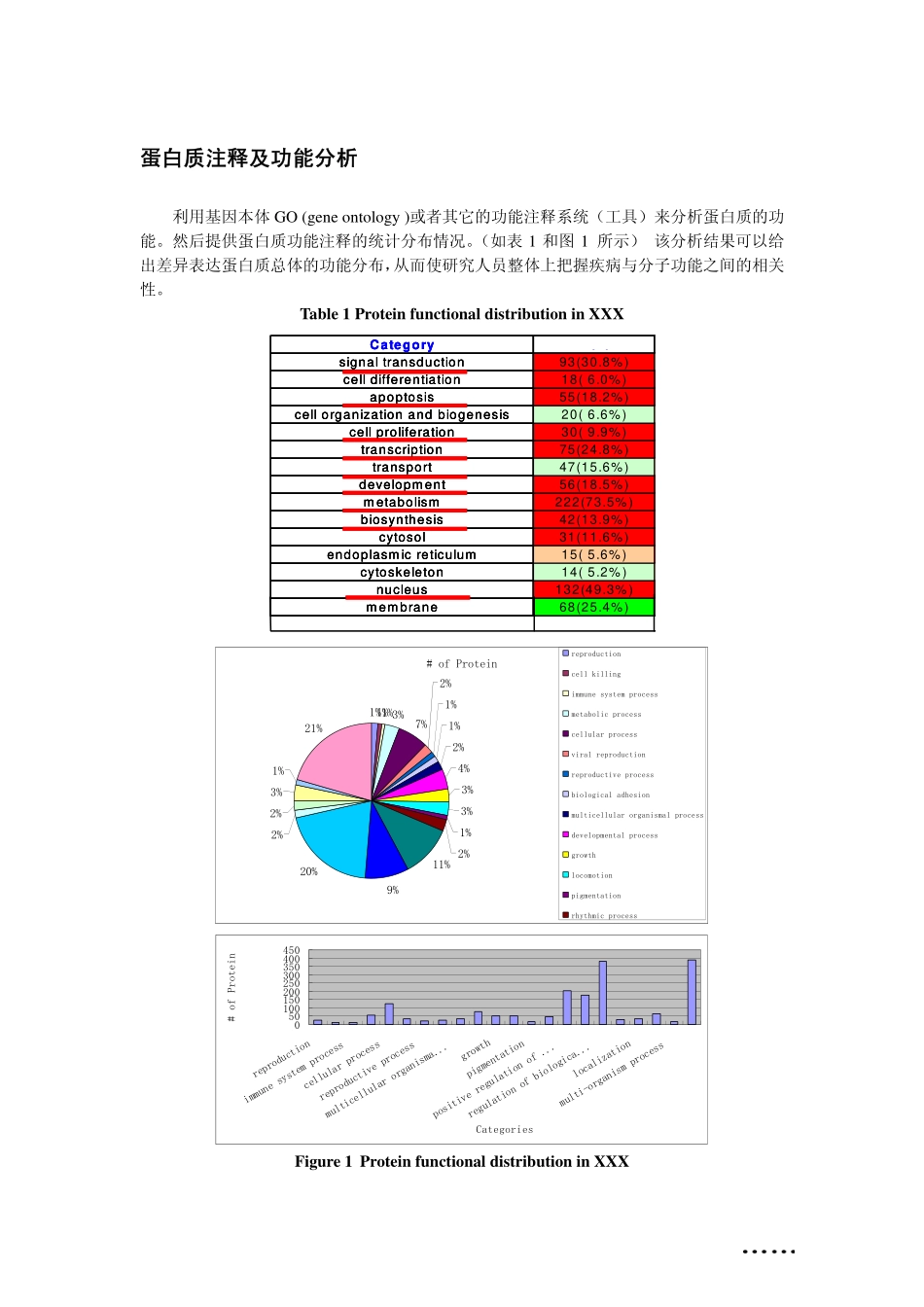
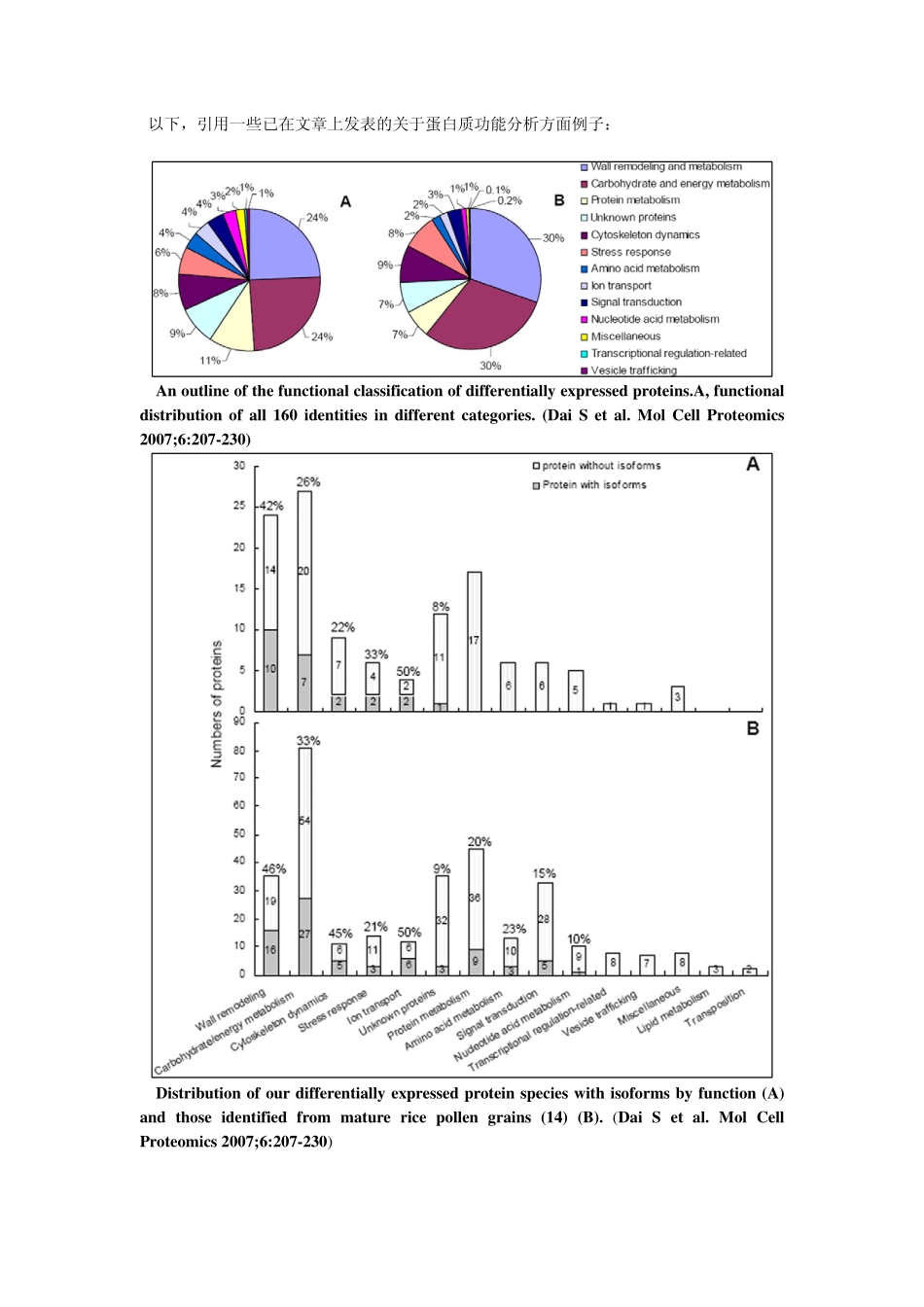
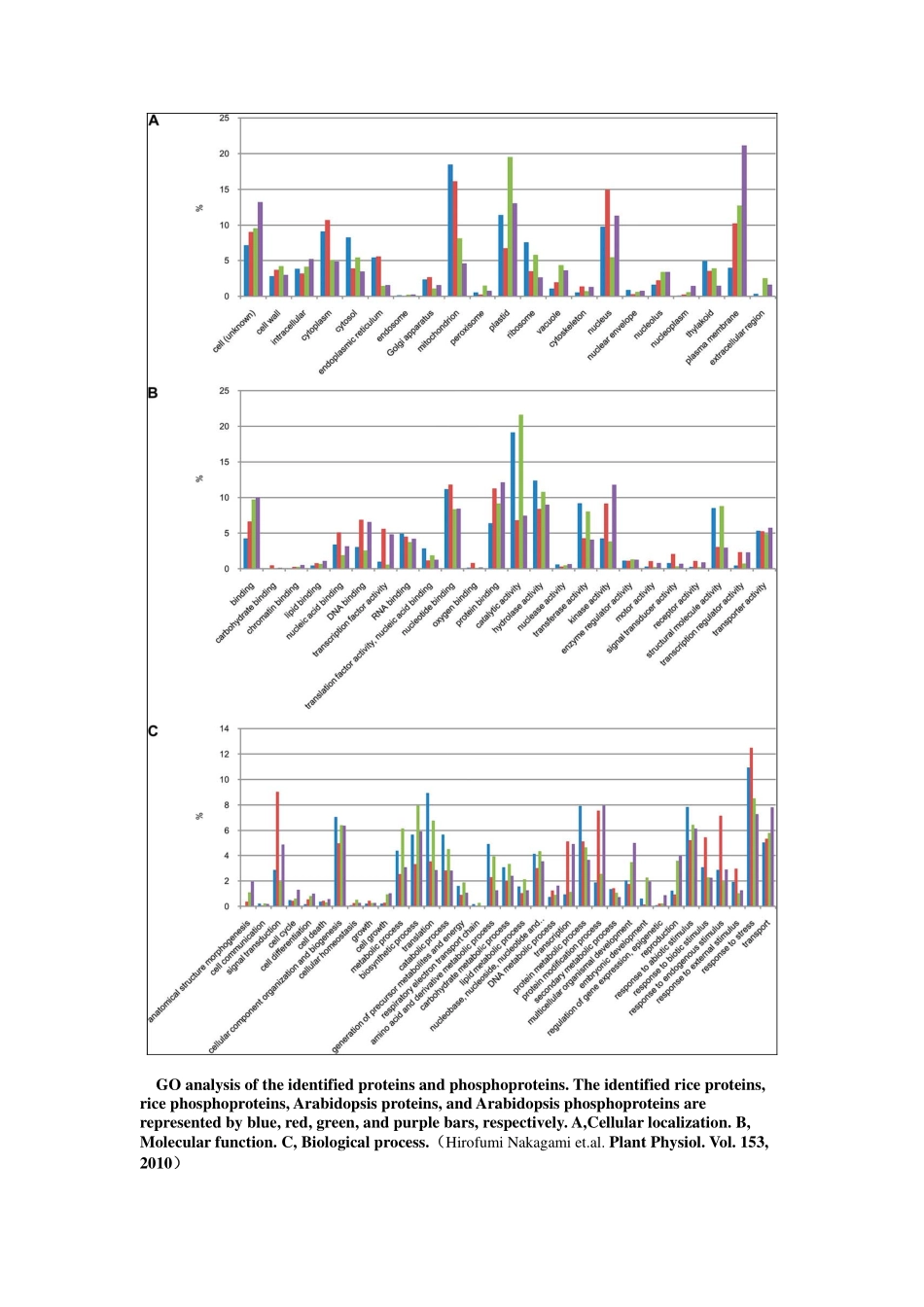
蛋白质注释及功能分析利用基因本体GO(geneontology)或者其它的功能注释系统(工具)来分析蛋白质的功能。然后提供蛋白质功能注释的统计分布情况。(如表1和图1所示)该分析结果可以给出差异表达蛋白质总体的功能分布,从而使研究人员整体上把握疾病与分子功能之间的相关性。Table1ProteinfunctionaldistributioninXXXCategoryBaitsignaltransduction93(30.8%)celldifferentiation18(6.0%)apoptosis55(18.2%)cellorganizationandbiogenesis20(6.6%)cellproliferation30(9.9%)transcription75(24.8%)transport47(15.6%)development56(18.5%)metabolism222(73.5%)biosynthesis42(13.9%)cytosol31(11.6%)endoplasmicreticulum15(5.6%)cytoskeleton14(5.2%)nucleus132(49.3%)membrane68(25.4%)CategoryBaitsignaltransduction93(30.8%)celldifferentiation18(6.0%)apoptosis55(18.2%)cellorganizationandbiogenesis20(6.6%)cellproliferation30(9.9%)transcription75(24.8%)transport47(15.6%)development56(18.5%)metabolism222(73.5%)biosynthesis42(13.9%)cytosol31(11.6%)endoplasmicreticulum15(5.6%)cytoskeleton14(5.2%)nucleus132(49.3%)membrane68(25.4%)CategoryBaitsignaltransduction93(30.8%)celldifferentiation18(6.0%)apoptosis55(18.2%)cellorganizationandbiogenesis20(6.6%)cellproliferation30(9.9%)transcription75(24.8%)transport47(15.6%)development56(18.5%)metabolism222(73.5%)biosynthesis42(13.9%)cytosol31(11.6%)endoplasmicreticulum15(5.6%)cytoskeleton14(5.2%)nucleus132(49.3%)membrane68(25.4%)#ofProtein1%1%1%3%7%2%1%1%2%4%3%3%1%2%11%9%20%2%2%3%1%21%reproductioncellkillingimmunesystemprocessmetabolicprocesscellularprocessviralreproductionreproductiveprocessbiologicaladhesionmulticellularorganismalprocessdevelopmentalprocessgrowthlocomotionpigmentationrhythmicprocess050100150200250300350400450reproductionimmunesystemprocesscellularprocessreproductiveprocessmulticellularorganisma...growthpigmentationpositiveregulationof...regulationofbiologica...localizationmulti-organismprocessCategories#ofProteinFigure1ProteinfunctionaldistributioninXXX以下,引用一些已在文章上发表的关于蛋白质功能分析方面例子:Anoutlineofthefunctionalclassificationofdifferentiallyexpressedproteins.A,functionaldistributionofall160identitiesindifferentcategories.(DaiSetal.MolCellProteomics2007;6:207-230)Distributionofourdifferentiallyexpressedproteinspecieswithisoformsbyfunction(A)andthoseidentifiedfrommaturericepollengrains(14)(B).(DaiSetal.MolCellProteomics2007;6:207-230)GOanalysisoftheidentifiedproteinsandphosphoproteins.Theidentifiedriceproteins,ricephosphoproteins,Arabidopsisproteins,andArabidopsisphosphoproteinsarerepresentedbyblue,red,green,andpurplebars,respectively.A,Cellularlocalization.B,Molecularfunction.C,Biologicalprocess.(HirofumiNakagamiet.al.PlantPhysiol.Vol.153,2010)附件信息:(如下图所示)在附件文件中,提供每个蛋白质不同层次的GO功能注释;以及各个层次GO功能所对应的蛋白质分布。蛋白质相互作用网络构建及分析(一)蛋白质相互作用构建及可视化蛋白质相互作用网络可视化展示,图中的网络节点可以编辑,直观地展示蛋白质间的相互作用情况,挑选重要蛋白。(A)(B)Figure2VisualizationofPPINetworkinXXXSupportingEvidencefortheProteinInteractions:IPI_AIPI_BProName_AProName_BScorePPI_DatabaseInterologInteracting_DomainGO_CoannotationGenome_ContextNetwork_TopologyIPI00003865IPI00012535HSPA8DNAJA1429446186.8VVVVVVIPI00292026IPI00386448CEBPDRELA1320.0XXVVXXIPI00032179IPI00300407SERPINC1SDC21.7e-001XXXXXXIPI00291578IPI00642461PRPSAP1PRPS21.0XXXXXXIPI00010320IPI00011857CBX1CHAF1B219.0VXXVXVIPI00020071IPI00215922NPM1NR2E3152.9XXVXXXIPI00000144IPI00218158OXTESR25.9e-001XXXXXXIPI00299399IPI00465084S100BDES454.2VXVVXVIPI00184584IPI00414264PSEN2SRI152.9XXVXXXIPI00010320IPI00031519CBX1DNMT1420911.2VXVVXVIPI00017292IPI00020070CTNNB1AR249.5XXVVXXIPI00006451IPI00336017NSFARRB15.9e-001XXXXXXIPI00006451IPI00031583NSFVDP719.7VXXVXVIPI00005040IPI00395811ACADMDAZL2.0e-001VXXXXX说明:除了给出蛋白质相互作用的可信度打分,还提供蛋白质相互作用在公共数据库中的信息、相互作用结构域、基因本体共注释等。StatisticsEvidenceInteractionsSubmittedHighConfLR>2.0PPI_DatabaseInterologInteracting_DomainGO_Coannotation#interaction1496177附件信息:在附件信息中详细标出每对蛋白质的相互作用可信度评分,这对蛋白质相互作用的相关数据库参考信息,相互作用结构域等等。(二)蛋白质相互作用网络深入分析(包括拓扑性质、相互作用结构域、重要节点以及网络模块化等)。下图展示了蛋白质相互作用网络模块以及模块间的crosstalk节点,这些节点相当于不同生物网络之间通讯的重要“关塞”,可以直观地显现其在药物靶标挖掘方面的潜力。Figure3Pathwaycrosstalksandfunctionalmoduls