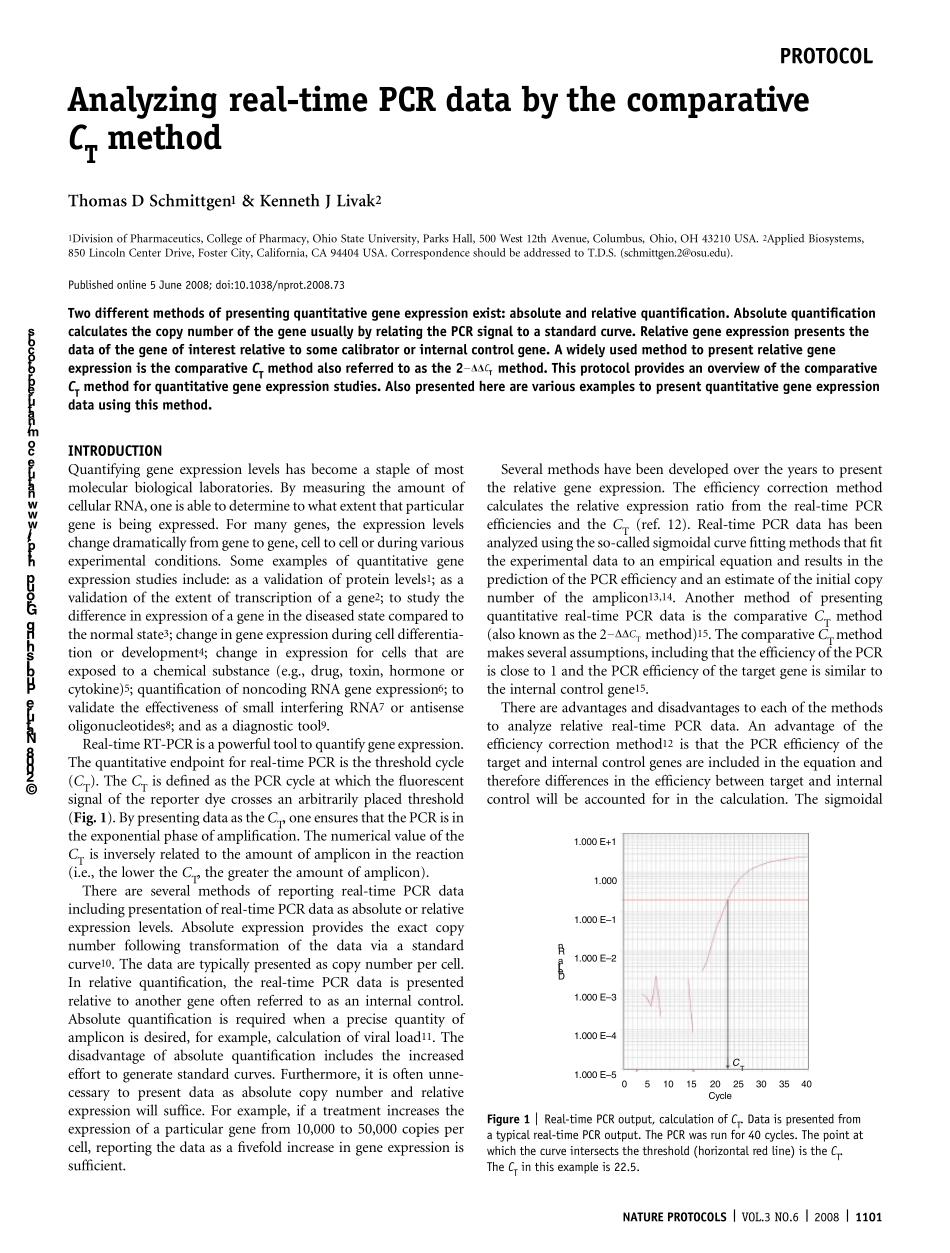
Analyzing real-time PCR data by the comparativeCT methodThomas D Schmittgen1 & Kenneth J Livak21Division of Pharmaceutics, College of Pharmacy, Ohio State University, Parks Hall, 500 West 12th Avenue, Columbus, Ohio, OH 43210 USA. 2Applied Biosystems,850 Lincoln Center Drive, Foster City, California, CA 94404 USA. Correspondence should be addressed to T.D.S. (schmittgen.2@osu.edu).Published online 5 June 2008; doi:10.1038/nprot.2008.73Two different methods of presenting quantitative gene expression exist: absolute and relative quantification. Absolute quantificationcalculates the copy number of the gene usually by relating the PCR signal to a standard curve. Relative gene expression presents thedata of the gene of interest relative to some calibrator or internal control gene. A widely used method to present relative geneexpression is the comparative CT method also referred to as the 2ÀDDCT method. This protocol provides an overview of the comparativeCT method for quantitative gene expression studies. Also presented here are various examples to present quantitative gene expressiondata using this method.INTRODUCTIONQuantifying gene expression levels has become a staple of mostmolecular biological laboratories. By measuring the amount ofcellular RNA, one is able to determine to what extent that particulargene is being expressed. For many genes, the expression levelschange dramatically from gene to gene, cell to cell or during variousexperimental conditions. Some examples of quantitative geneexpression studies include: as a validation of protein levels1; as avalidation of the extent of transcription of a gene2; to study thedifference in expression of a gene in the diseased state compared tothe normal state3; change in gene expre...